| Saffron is an amphiploid with probable low genetic diversity
One important aspect of our issue is to ascertain the genetic diversity of C. sativus . Saffron usually multiplies year by year by means of corms. Because corm multiplication does not induce genome variations with the exception of some mutation that in a triploid saffron population are not easily detectable, all saffron should be similar one to the other. Some authors believed that saffron was once naturalized in small areas from where it was lost as a consequence of a change in land use. C. sativus was generally assumed to be of autotriploid or hybrid origin. Now we have several data that support the alloploidy of C. sativus being the parents C. cartwrightianus and C. hadriaticus, both with 2n=16 and present currently in Greece. Other possible parents, e.g., C. thomasi, from Italy and Croatia, C. mathewii from Turkey, and C. pallasii ssp. haussknechtii from Iran-Iraq-Jordan, cannot be excluded. The complexity of the evolutionary history of the genus Crocus suggests an intensive species hybridisation and explosive speciation in Crocus evolution that could be on the basis of the origin of saffron. We now are sure that saffron is an allopolyploid but the localization of the hybridisation event has not been ascertained so far. If the event took part several times could have generated different amphiploids and, in consequence, different saffron lines. Saffron was introduced in Western Europe in different historical moments: In the Iberian Peninsula by the Arabs or even before (Romans); in Germany, Switzerland, France, Italy and Great Britain by the Crusaders, and towards the east it was extended by a variety of cultures through Transcaucasia, China, India and eventually Japan. Should we expect to find genetic diversity then? |
|
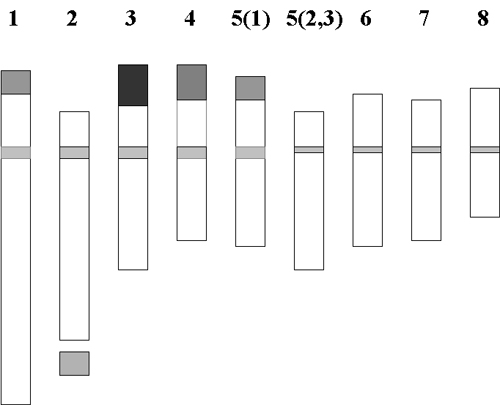
We can look the chromosomes as a first approach. The karyotype of C. sativus has been studied by a number of authors, reporting that saffron from different countries (Azerbaijan , Iran , Italy , Turkey , France and England) was always 2n=3x=24 lacking karyologycal differences. However in the past literature other karyotypes were described. Cytofluorimetric analysis on nuclear DNA, carried to detect genome size and base pairs composition, revealed no differences in DNA content and composition in saffron corms from different countries (Italy , Israel , Spain , Holland). S ome assays with molecular markers (RAPDs) have revealed limited genetic differences among saffron samples from Italy , Iran , Greece and Spain . Nevertheless, analysis of phenotype revealed differences in aspect flower size, tepal shape and colour intensity with lobed tepal in plants from Israel and more intense colour of tepals in plants from Sardinia (Italy). Variants of saffron with an increased number of stigmas, maintaining 2n=24 have been reported with a frequency of 1.2x10 -6 of the rare type flowers. Such phenotypes are well known by farmers and breeders but unfortunately they are not stable. Morphological differences with flowers having higher number of style branches and stamens have been already described in saffron cultivation at L'Aquila (Italy). Phenotypic variants are also present in Kashmir saffron (F. Nehvi, personal communication). |
Besides different commercial products are known that could suggest the existence of different saffron ecotypes or commercial varieties, the actual genetic variability present in C. sativus at worldwide scale is currently unknown. Nevertheless, there is a suspicion in saffron breeders regarding the existence of scarce genetic variability in this crop, but no serious effort has been carried out to ascertain this important issue. They have been attempts with the objective to increase the spectrum of variability for floral traits and recovering auto-hexaploids (6x=48) in saffron (to break the sterility barrier) by colchinization and to induce the genetic variability in saffron using physical irradiation in order to develop polyploid forms; tetrafid, pentafid or hexafid stigmatic plants; and colour mutants. These attempts have been unsuccessful so far (for review see Fernández, J.A. Recent. Res. Devel. Plant Sci., 2: 127-159). |